Co-evolution in microbial communities
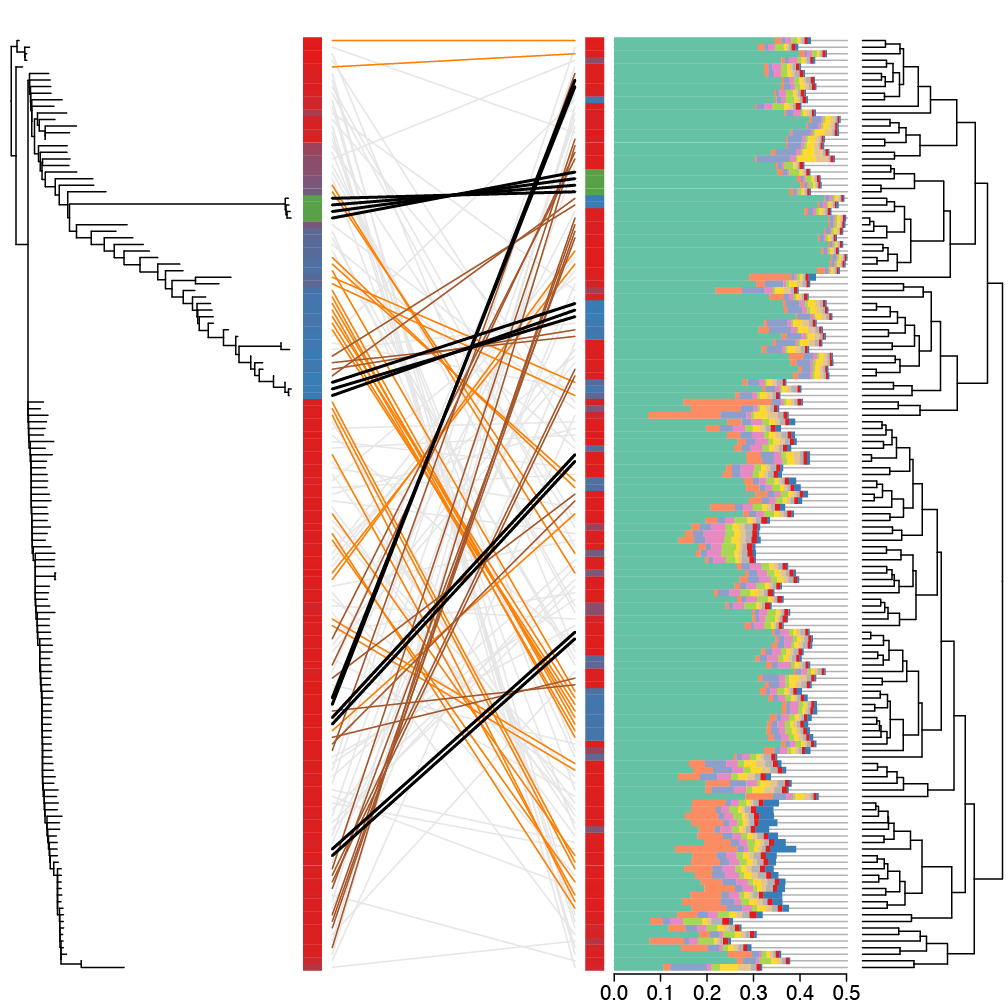
Species communities in ecosystem are mostly studied with respect to their overal community structure, and in particular how the interactions between species influence the stability and function of the community. The type and structure of these interactions, however, will also affect the evolution of the individual members within a community. This is particularly important for microbial communities, where evolutionary and ecological processes happen on similar timescales. I am interested in how interactions between microbes within consortia influence not only the functioning of the communitiy, but also the evolutionary outcomes that are expected when these communities are exposed to abiotic environmental changes. This approach goes beyond the current trends in the field microbial ecology and integrates evolutionary considerations into the study marine communities, industrial bioreactor communities and human/animal microbiomes.